Building on the extensive mult-omics and brain imaging datasets, we focus on the development and application of multi-omic computational models aimed at 1) exploring the functional mechanisms linked to Alzheimer risk variants (i.e., functional genomics) and 2) modeling disease progression to improve early detection and risk assessment. Our lab stands in between biomedical and AI research community, performing highly interdisciplinary research from computational modeling to investigation of specific disease problems.
Functional genomics
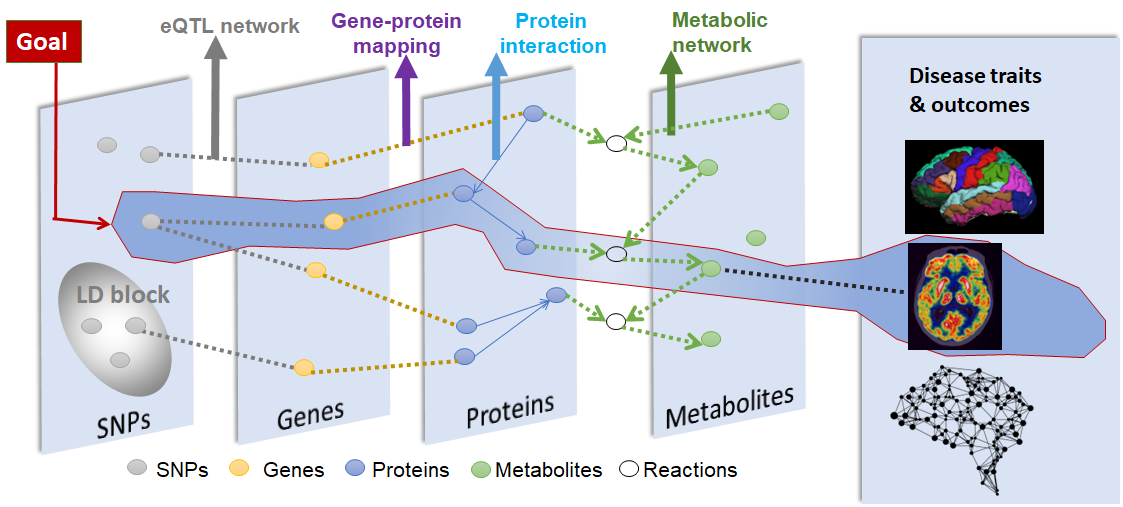 Genome wide association studies has revealed a set of significant variants associated with Alzheimer’s disease. However, the downstream biology through which they exert effect on the development of AD is largely unknown. While multi-omics data offers multi-scale perspective of disease, one major challenge remains as how to relate the significant findings from different -omics types, aiming for a complete view of interplays between -omics layers that contribute to AD development. Toward that, we develop computational models that leverage the prior biological interaction networks as an additional source of evidence on top of multi-omics data. We aim to reveal functional units rather than individual SNP/gene/proteins associated with disease outcomes. Our recent interest lies in the applications of our methods in more targeted molecular mechanisms like multi-enhancer interactions and in the fair modeling for inclusion of minority groups.
Genome wide association studies has revealed a set of significant variants associated with Alzheimer’s disease. However, the downstream biology through which they exert effect on the development of AD is largely unknown. While multi-omics data offers multi-scale perspective of disease, one major challenge remains as how to relate the significant findings from different -omics types, aiming for a complete view of interplays between -omics layers that contribute to AD development. Toward that, we develop computational models that leverage the prior biological interaction networks as an additional source of evidence on top of multi-omics data. We aim to reveal functional units rather than individual SNP/gene/proteins associated with disease outcomes. Our recent interest lies in the applications of our methods in more targeted molecular mechanisms like multi-enhancer interactions and in the fair modeling for inclusion of minority groups.
Disease Progression
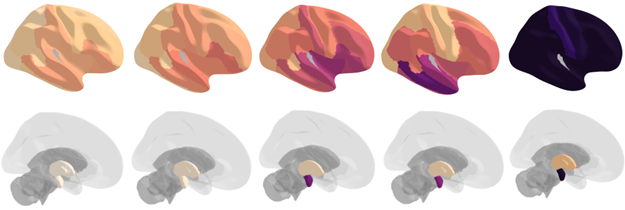 Alzheimer’s Disease is an irreversible neurodegenerative disorder with a long prodromal phase and no clinically validated cure. Detecting when and how molecular and imaging phenotype marker develop along AD progression will provide a template for understanding the underlying etiology of clinical syndromes and for improving early diagnosis, clinical trial recruitment and treatment assessment. We develop and apply interpretable AI approaches to profile, subtyping and temporally align the progression patterns across molecular -omics and imaging modalities.
Alzheimer’s Disease is an irreversible neurodegenerative disorder with a long prodromal phase and no clinically validated cure. Detecting when and how molecular and imaging phenotype marker develop along AD progression will provide a template for understanding the underlying etiology of clinical syndromes and for improving early diagnosis, clinical trial recruitment and treatment assessment. We develop and apply interpretable AI approaches to profile, subtyping and temporally align the progression patterns across molecular -omics and imaging modalities.
Strategies for imbalanced, incomplete and heterogeneous multi-omic data.
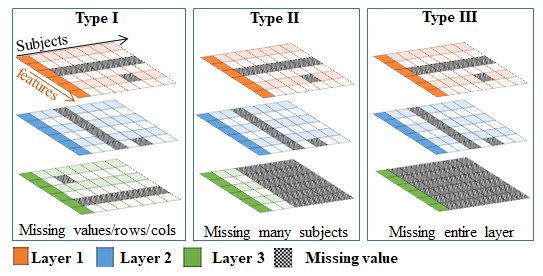 Despite the great potential of multi-omics data, they are mostly imbalanced (in sex, age and racial groups), incomplete (missing data in -omics types), and of heterogeneous types (e.g., continuous and categorical numbers). Integrating these data for joint analysis typically requires exclusion of many subjects with missing values or biased results with poor generalizability. Toward this direction, we develop novel computational models for functional genomics and disease progression analysis which specifically tackle these challenging problems in multi-omics data and hereafter allow biomedical researchers to gain more insights from rapidly growing yet imperfect biomedical data.
Despite the great potential of multi-omics data, they are mostly imbalanced (in sex, age and racial groups), incomplete (missing data in -omics types), and of heterogeneous types (e.g., continuous and categorical numbers). Integrating these data for joint analysis typically requires exclusion of many subjects with missing values or biased results with poor generalizability. Toward this direction, we develop novel computational models for functional genomics and disease progression analysis which specifically tackle these challenging problems in multi-omics data and hereafter allow biomedical researchers to gain more insights from rapidly growing yet imperfect biomedical data.